News
Updated chromosome-scale CH assembly and CH/CHO-K1 annotations
January 27, 2021
A significantly more continuous version of the Chinese hamster genome, CH PICRH (GCF_003668045.3), along with its 2020 RefSeq annotation is now on CHOgenome.org. Long-range scaffolding of the previous CH genome assembly was performed using high-throughput chromosome conformation capture (Hi-C) and now 97% of the genome is contained in 11 large scaffolds corresponding to the CH chromosomes. In addition, the updated 2020 RefSeq annotation for the CHO-K1 cell line is available. Both can be searched (gene search and BLAST) and viewed on JBrowse. More information on the most recent version of the Chinese hamster genome can be found here.
Updated CH assembly and CH/CHO-K1 annotations
May 1, 2019
We are excited to announce that the significantly improved Chinese hamster genome, CH PICR (GCF_00366804.1), along with its 2018 RefSeq annotation is now online. In addition, an updated RefSeq annotation for the CHO-K1 cell line is now available. Both can be searched (gene search and BLAST) and viewed on JBrowse. More information on the updated Chinese hamster genome can be found here.
New mRNA Expression Browser (Beta)
June 7, 2017
The browser (beta version) for the visualization of mRNA expression from CHO-K1 cells is now online. The data is from several published DNA-microarray or RNA-Seq experiments. The tutorial on how to use this browser can be found here.
Tutorial 3 – Searching the Chinese Hamster Genome Database
August 2014, Version 2.0
Click here for PDF version.
Multiple Chinese hamster (CH) and CHO-K1 genome search pages are hosted on the CHO genome website. A comprehensive search function for the RefSeq CH and CHO-K1 genomes is located on the homepage. Additionally, an independent search page with additional selection details for the RefSeq genomes is available under the Genomes menu. Separate search pages for both the CH mitochondrial and GenBank CHO-K1 genomes are also available under the Genomes tab. The comprehensive RefSeq search function on the homepage can be accessed from any CHO genome webpage by selecting the CHO GENOME logo located in the top left corner.
Searching the RefSeq Chinese hamster (CH) or CHO-K1 Genomes
As of August 2014, there are 3 CH and CHO-K1 RefSeq genome databases. The initial CHO RefSeq genome that was hosted on CHOgenome.org is the CHO-K1 RefSeq (2012) genome database. The genomic information in this genome database has not been altered since its release in 2012, so it is not aligned with the most recent CHO-K1 genome database. The CHO-K1 RefSeq (2012) genome database is the database to search if you have previously used the CHO-K1 RefSeq (2012) database and are interested in obtaining identical results. In May 2014, NCBI released their first complete CH genome database and an updated CHO-K1 genome database. Currently, these two databases are the most recent CH and CHO genome databases to search against.
1) The comprehensive search function for the RefSeq CH and CHO-K1 genomes is located on the CHO genome homepage.
The gene name, gene symbol, or NCBI gene ID can be searched against all of the RefSeq CH and CHO genomes. After entering the search term and selecting the "Search" button, all relevant results are displayed.
2) To specify the search terms or RefSeq genomes to be searched, select the CH & CHO RefSeq link from the Genomes menu, the Advanced Search button on the homepage, or the Gene Search image button located on the homepage.
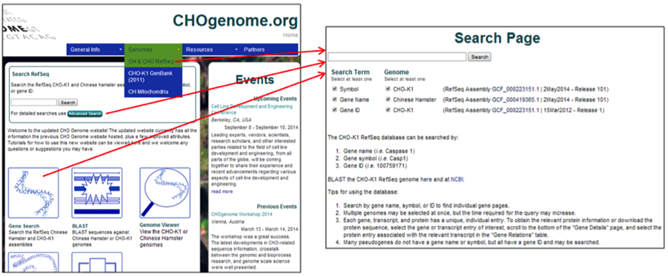
Select the type of keyword and genome(s) you would like to search, type the keyword into the search box, and press the Search button or hit enter.
The RefSeq genome databases can be searched by:
Gene symbol (i.e. cfl1)
The database can be searched using the official gene symbols assigned to gene products during the NCBI genome annotation projects. Unofficial gene symbols are not searchable and if the gene was not assigned a gene symbol, one was created. The created gene symbols consist of 'LOC' followed by the 9-digit NCBI gene ID (i.e. LOC100######).

Gene name (i.e. cofilin 1)
The database can be searched using the protein names assigned to the genes during the genome annotation projects.

NCBI Gene ID (i.e. 100760435)
The database can be searched using the NCBI gene IDs. For the CHO-K1 and CH genomes, these gene IDs currently range between 100682525 and 103163833.
All CHO and Chinese hamster NCBI gene IDs can be found in the NCBI protein database by using the search term "Cricetulus griseus."

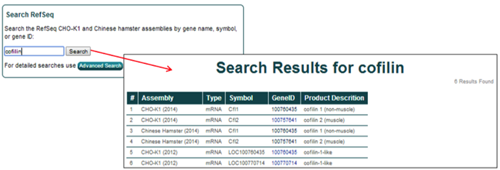
Matches for the genome database searches are displayed in a tabular format. Searching for cofilin in the CHO-K1 (2014), Chinese Hamster (2014), and CHO-K1 (2012) assemblies returns 6 entries.
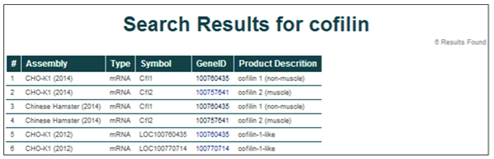
The general characteristics displayed for the search results include the parent assembly, gene feature type, gene symbol, NCBI gene ID, and gene name/product description.
To access more details about a single entry, click on the NCBI gene ID [i.e. 100757641].
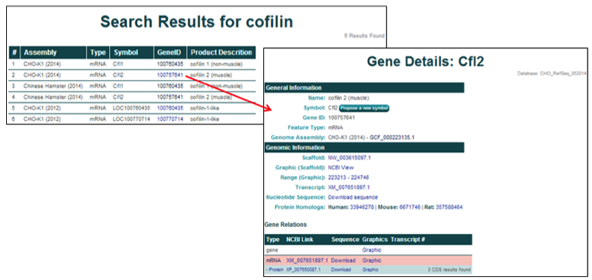
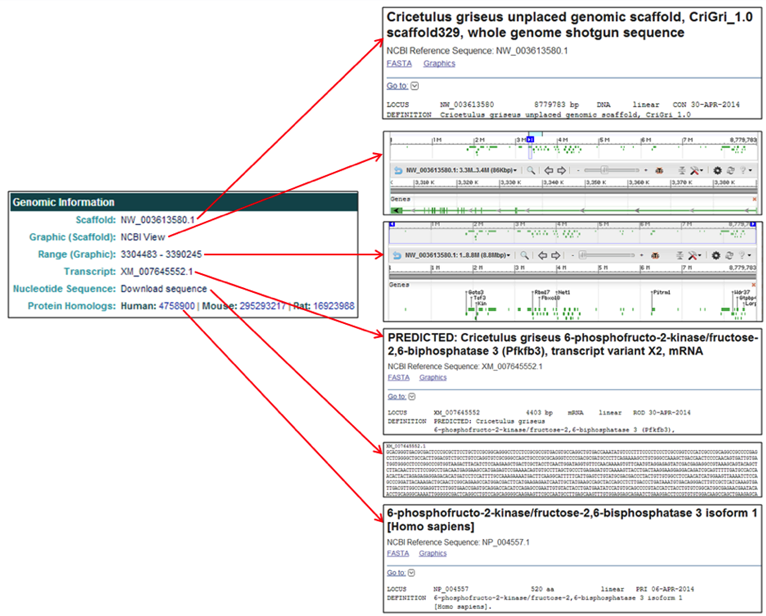
3) Searching and selecting the NCBI gene ID 100762756 from the CHO-K1 (2014) genome assembly will open the 'Gene Details' page for Pfkfb3, Variant X2.
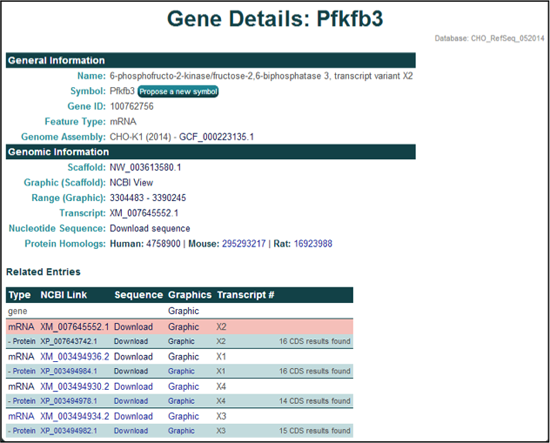 The General Information section provides the gene name, gene symbol, NCBI gene ID, feature type, and the genome assembly of origin.
The General Information section provides the gene name, gene symbol, NCBI gene ID, feature type, and the genome assembly of origin.
The Genomic Location section provides the scaffold on which the gene is located, the coordinates of the gene's coding region, the NCBI transcript ID, a link to the NCBI graphics page, a link to the FASTA nucleotide or amino acid sequences, and links to the NCBI protein homologs in human, mouse, and rat.
The Related Entries section provides information and links to all of the features associated with each gene, including all of the relevant transcripts and proteins, in one table. Each row represents one feature associated with the gene and the highlighted row identifies the feature thats content is displayed on the current page. The table columns contain the following information:
- Type lists the row's feature type.
- NCBI Link displays the NCBI transcript/protein IDs.
- The Sequence Download links to the FASTA nucleotide or protein sequence.
- The Graphics Graphic links to the NCBI Map Viewer graphic associated with the row's gene feature type.
- Transcript # contains the NCBI transcript # if multiple transcripts exist for the gene of interest.
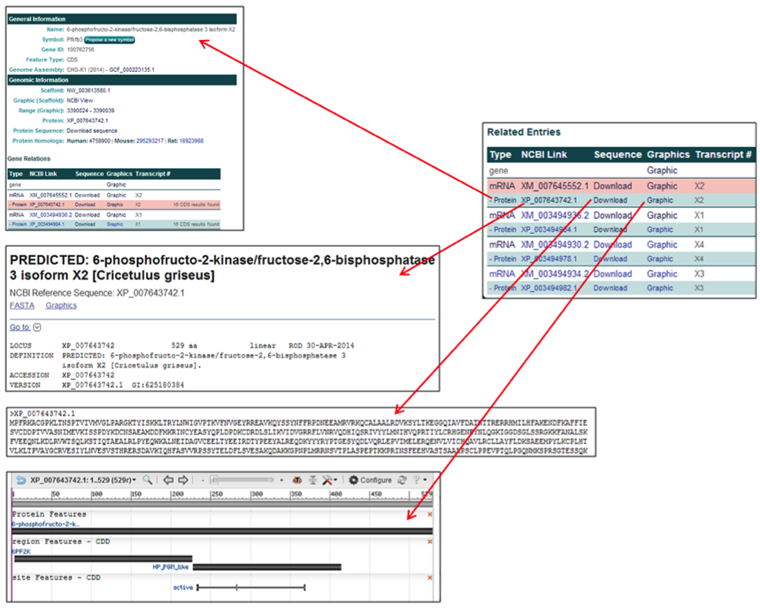
4) All hyperlinked text serves as a link to a corresponding webpage. Any green button with white text located on the 'Gene Details' webpage enables direct communication with the editors of CHO genome. These webpages are opened in a new tab, conserving the user's current 'Gene Details' webpage.
The General Information section contains one button and one hyperlink.
- The Propose a new symbol button allows the user to propose a new/different gene symbol with an accompanying reason for the proposed change. These submissions are cataloged and periodically evaluated.
- The Genome Assembly link [i.e. GCF_000223135.1], opens the webpage of the NCBI genome assembly associated with the selected gene.
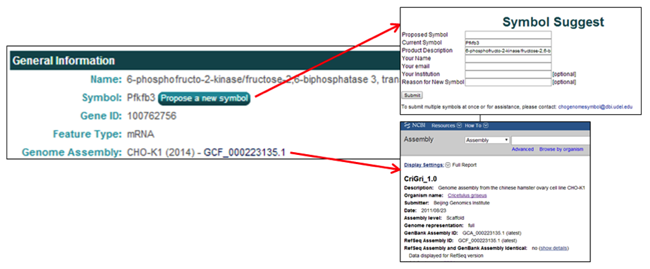
The Genomic Location section provides links to multiple pages related to the genomic location information for the selected gene.
- The Scaffold link [i.e. NW_003613580.1] links to the NCBI scaffold information page of the scaffold containing the selected gene.
- The Graphic (Scaffold) link [NCBI View] links to the full NCBI Genome Viewer image of the scaffold containing the selected gene.
- The Range (Graphic) link [i.e. 3304483-3390245] links to the NCBI Genome Viewer image zoomed in to the selected gene.
- The Transcript link [i.e. XM_007645552.1] links to the NCBI mRNA information page for the selected gene.
- The Nucleotide Sequence link [Download sequence] opens a page with the selected gene's mRNA FASTA file.
- The Protein Homologs link [i.e. Human: 4758900] lists the NCBI protein homolog IDs of the human, mouse, and rat homologs and link to the selected gene's NCBI protein webpage.
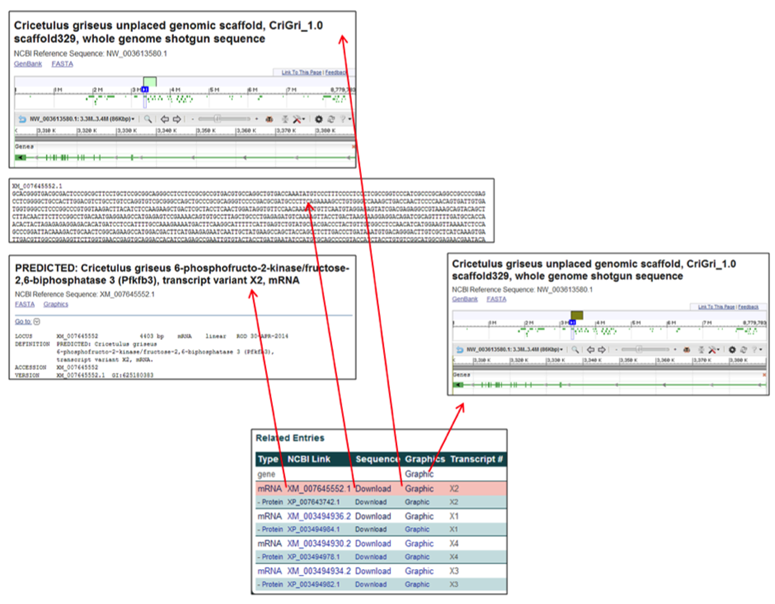
The Related Entries table provides links to the Chinese hamster genome pages, gene sequences, NCBI database information, and NCBI graphics pages.
The gene & RNA entries contain links to relevant webpages associated with the nucleotide entries.
- The NCBI Link [i.e. XM_007645552.1] links to the NCBI RNA-specific information page corresponding to the selected gene.
- The Sequence link [Download] provides the nucleotide FASTA file for the corresponding gene transcript.
- The Graphics link [Graphic] links to the NCBI Genome Viewer image of the selected gene's scaffold (for a gene row entry) or of the zoomed-in, transcript-specific portion of the scaffold (for a RNA row entry).
The protein row entries contain links to relevant webpages associated with the amino acid entries.
Note: The protein entry following each RNA entry is the corresponding protein, as often indicated by the transcript #.
- The Type link [Protein] links to the CHO genome results page associated with that protein entry.
- The NCBI Link [i.e. XP_007643742.1] links to the NCBI information page for the selected protein.
- The Sequence link [Download] provides the amino acid FASTA file for the selected protein.
- The Graphics link [Graphic] links to the zoomed NCBI Genome Viewer image of the selected protein.
Searching the CHO-K1 GenBank (2011) Genome
1) Select the CHO-K1 GenBank (2011) genome from the Genomes menu.
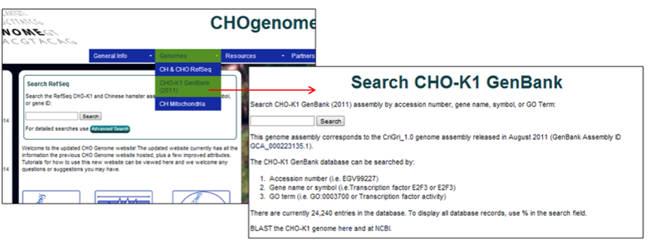
The CHO-K1 GenBank (2011) genome database is the initial genome database that was hosted on CHOgenome.org. The genome information does not align with the most recent CHO-K1 genome information, as the content has not been altered since its release in 2011. Selection of the CHO-K1 GenBank (2011) genome database is advantageous if the objective is the replication of previous results from the CHO-K1 GenBank (2011) genome database.
2) To search this database, type a keyword into the search box at the top of the page and submit your query. Typing % in the search box will list all 24,240 entries currently in the database.
The CHO-K1 genome database can be searched by:
Accession number (i.e. EGV99227)
The database can be searched using the GenBank WGS protein accession IDs. For the CHO-K1 WGS project, these accession numbers are EGV##### or EGW#####, where # is any digit 0-9.
These accession numbers can also be found in the NCBI protein database by restricting the search to "Cricetulus griseus" and the locus tag to "I79_######."
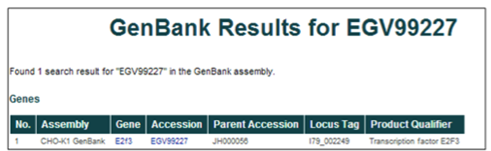
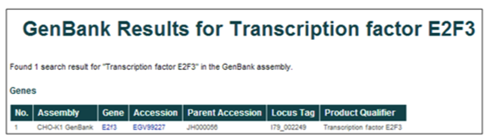
Gene name or gene symbol (i.e. Transcription factor E2F3 or E2F3)
The database can be searched using gene names or gene symbols. The gene names were assigned to gene products during the annotation of the WGS project, while the gene symbols were assigned based on the annotation of homologous proteins.
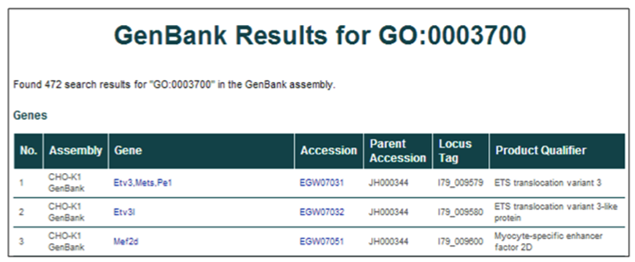
Gene ontology (GO) term (i.e. GO:0003700 or Transcription factor activity)
The database can be searched using a GO accession ID or term. Searching by GO accession ID or term will return all database entries annotated with that GO term.
3) The results from the GenBank searches are listed with 6 identifying characteristics per row.
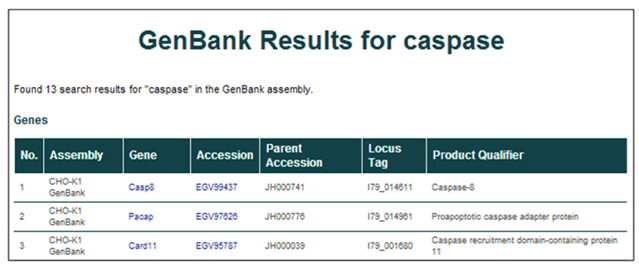
The returned entries' characteristics include the assembly of origin (CHO-K1 GenBank), gene symbol, protein accession ID, scaffold (parent accession), locus tag, and gene name/description (product qualifier).
4) To find more details about a single entry, select the gene symbol or accession ID [i.e. Casp8 or EGV99437].
 The General Information section provides the gene name, symbols, synonyms, locus tag, descriptions, and assembly of origin.
The General Information section provides the gene name, symbols, synonyms, locus tag, descriptions, and assembly of origin.
The Genomic Information section provides the WGS scaffold on which the gene is located, the coordinates of the gene coding region, and links to graphics of the scaffold and the gene.
The Sequence Information section provides links to download the nucleotide and protein sequence in FASTA format.
The External Links section provides links to the corresponding CHO protein entries in the NCBI and EMBL databases.
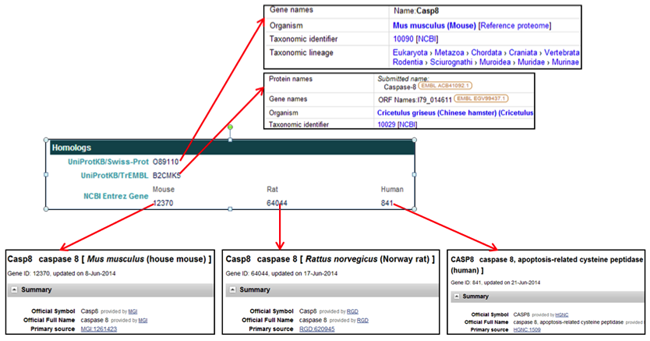
The Homologs section provides links to homologous proteins in other species in the UniProt databases and to homologous proteins in mouse, rat, and human in the NCBI Entrez databases.
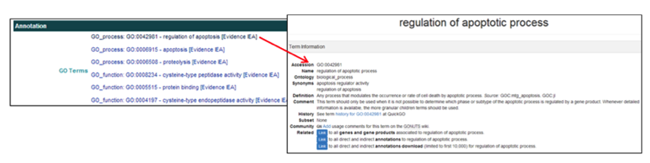
The Annotation section provides functional information and GO terms assigned to the gene product. The GO terms link to the AmiGO gene ontology browser.
5) Hyperlinked text also serves as a link to additional relevant webpages.
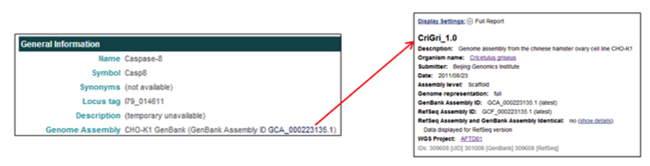
The General Information section provides a link associated with the WGS assembly.
- In the Genome Assembly field, clicking on the genome assembly ID [i.e. GCA_000223135.1] opens the corresponding NCBI assembly report the selected gene was annotated from.
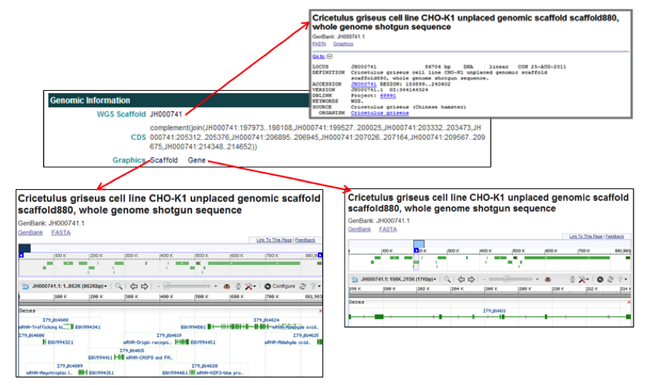
The Genomic Information section provides links associated with the WGS scaffold on which the selected gene is located and graphics of the scaffold and gene.
- In the WGS Scaffold field, clicking on the scaffold accession ID [i.e. JH000741] opens the NCBI database entry for the WGS scaffold the selected gene is located on.
- In the Graphics field, clicking on Scaffold opens the NCBI Map Viewer to view the genome annotation of the entire scaffold, while clicking on Gene opens the NCBI Map Viewer to view the genome annotation of the selected gene model portion of the scaffold.
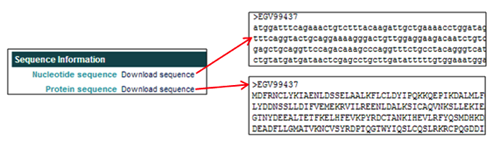
The Sequence Information section provides links to download the nucleotide and protein sequences in FASTA format.
- In the Nucleotide sequence field, clicking on Download sequence opens the nucleotide FASTA file.
- In the Protein sequence field, clicking on Download sequence opens the amino acid FASTA file.
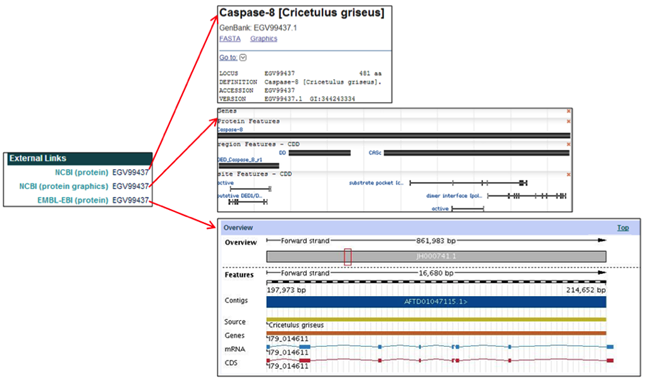
The External Links section provides links to the selected CHO protein pages in the NCBI and EMBL databases.
- In the NCBI (protein) field, clicking on the protein accession ID [i.e. EGV99437] opens the NCBI webpage of the selected protein entry.
- In the NCBI (protein graphics) field, clicking on the protein accession ID [i.e. EGV99437] opens the NCBI Map Viewer and visually shows the selected protein, region, and site features.
- In the EMBL-EBI (protein) field, clicking on the protein accession ID [i.e. EGV99437] opens the EMBL-EBI webpage containing both a visual and informational overview of the selected protein.
The Homologs section provides links to homologs of the selected CHO gene in the NCBI and UniProt databases.
- In the UniProtKB/Swiss-Prot field, clicking on the protein accession ID [i.e. OB9110] opens the UniProtKB/Swiss-Prot webpage displaying the information for a homolog to the CHO protein.
- In the UniProtKB/TrEMBL field, clicking on the protein accession ID [i.e. B2CMK5] opens the UniProtKB/TrEMBL webpage displaying the information for a homolog to the CHO protein.
- In the NCBI Entrez Gene field, clicking on the Mouse protein accession ID [i.e. 12370] opens the NCBI gene webpage for the mouse homolog to the CHO protein.
- In the NCBI Entrez Gene field, clicking on the Rat protein accession ID [i.e. 64044] opens a NCBI gene webpage for the rat homolog to the CHO protein.
- In the NCBI Entrez Gene field, clicking on the Human protein accession ID [i.e. 841] opens the NCBI gene webpage for the human homolog to the CHO protein.
The Annotation section provides links to the GO Terms in the AmiGO databases.
- In the GO Terms field, clicking on the GO Term ID [i.e. GO_Process: GO:0042981] opens the AmiGO webpage displaying the description and characteristics of the GO Term.
Searching the Chinese hamster mitochondrial genome
1) Select the Chinese hamster mitochondrial genome from the Genomes menu [i.e. CH Mitochondria].
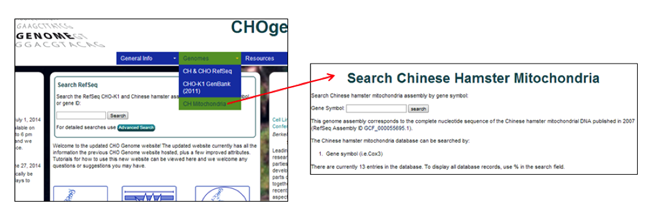
2) Type a keyword into the search box at the top of the page.
Typing % in the search box will list all 13 entries currently in the database.
The Chinese hamster mitochondrial genome database can only be searched by Gene symbol [i.e. COX].
Searching for "COX" will return 3 entries from the database displayed in a tabular format.
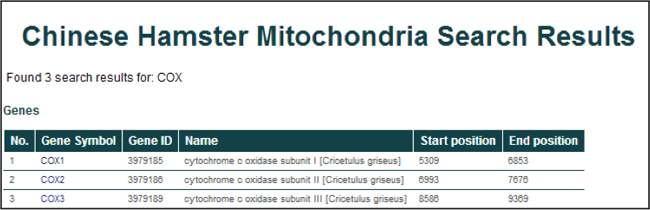
General information for the returned entries include the gene symbol, NCBI Entrez Gene ID, gene name, and genomic coordinates (start and end positions).
To find more details about a single entry, click on a gene symbol.
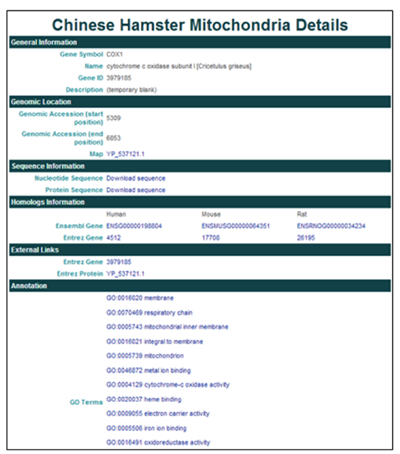 3) Clicking on COX1 will open the Gene Details page.
3) Clicking on COX1 will open the Gene Details page.
The General Information section contains the gene name, symbol, ID, and description (if available).
The Genomic Information section provides the genomic coordinates in the mitochondrial genome and Entrez protein ID.
The Sequence Information section provides links to download the nucleotide and protein sequence in FASTA format.
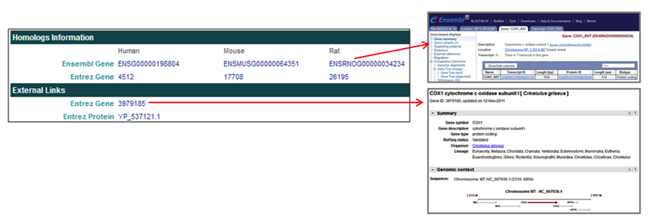
The Homologs Information section provides links to homologous proteins in other species in the NCBI and Ensembl databases.
The External Links section provides links to CHO gene and protein entries in the NCBI databases.
The Annotation section reports GO terms and provides links to the AmiGO gene ontology browser.
4) Hyperlinked text also serves as a link to relevant informational webpages.
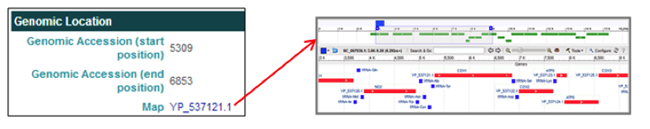
The Genomic Location section Map entry [i.e. YP_537121.1] links to the NCBI Map Viewer to view the selected gene's annotation within the mitochondrial genome.
The Sequence Information section provides Nucleotide Sequence and Protein Sequence links [i.e. Download sequence] to download the nucleotide and protein sequences in FASTA format.

The Homologs Information and External Links sections provide links to CHO proteins and homologs.
- The Homologs Information section provides Ensembl Gene and Entrez Gene links
[i.e. ENSRNOG000000034234] to the Ensembl and NCBI Entrez databases for CHO proteins and homologs in other species, including human, mouse, and rat. - The External Links section provides Entrez Gene and Entrez Protein links [i.e. 3979185] to CHO gene and protein pages in the NCBI Entrez database.