News
Updated chromosome-scale CH assembly and CH/CHO-K1 annotations
January 27, 2021
A significantly more continuous version of the Chinese hamster genome, CH PICRH (GCF_003668045.3), along with its 2020 RefSeq annotation is now on CHOgenome.org. Long-range scaffolding of the previous CH genome assembly was performed using high-throughput chromosome conformation capture (Hi-C) and now 97% of the genome is contained in 11 large scaffolds corresponding to the CH chromosomes. In addition, the updated 2020 RefSeq annotation for the CHO-K1 cell line is available. Both can be searched (gene search and BLAST) and viewed on JBrowse. More information on the most recent version of the Chinese hamster genome can be found here.
Updated CH assembly and CH/CHO-K1 annotations
May 1, 2019
We are excited to announce that the significantly improved Chinese hamster genome, CH PICR (GCF_00366804.1), along with its 2018 RefSeq annotation is now online. In addition, an updated RefSeq annotation for the CHO-K1 cell line is now available. Both can be searched (gene search and BLAST) and viewed on JBrowse. More information on the updated Chinese hamster genome can be found here.
New mRNA Expression Browser (Beta)
June 7, 2017
The browser (beta version) for the visualization of mRNA expression from CHO-K1 cells is now online. The data is from several published DNA-microarray or RNA-Seq experiments. The tutorial on how to use this browser can be found here.
CHO-K1 reference map
Click on a quadrant to view proteins.
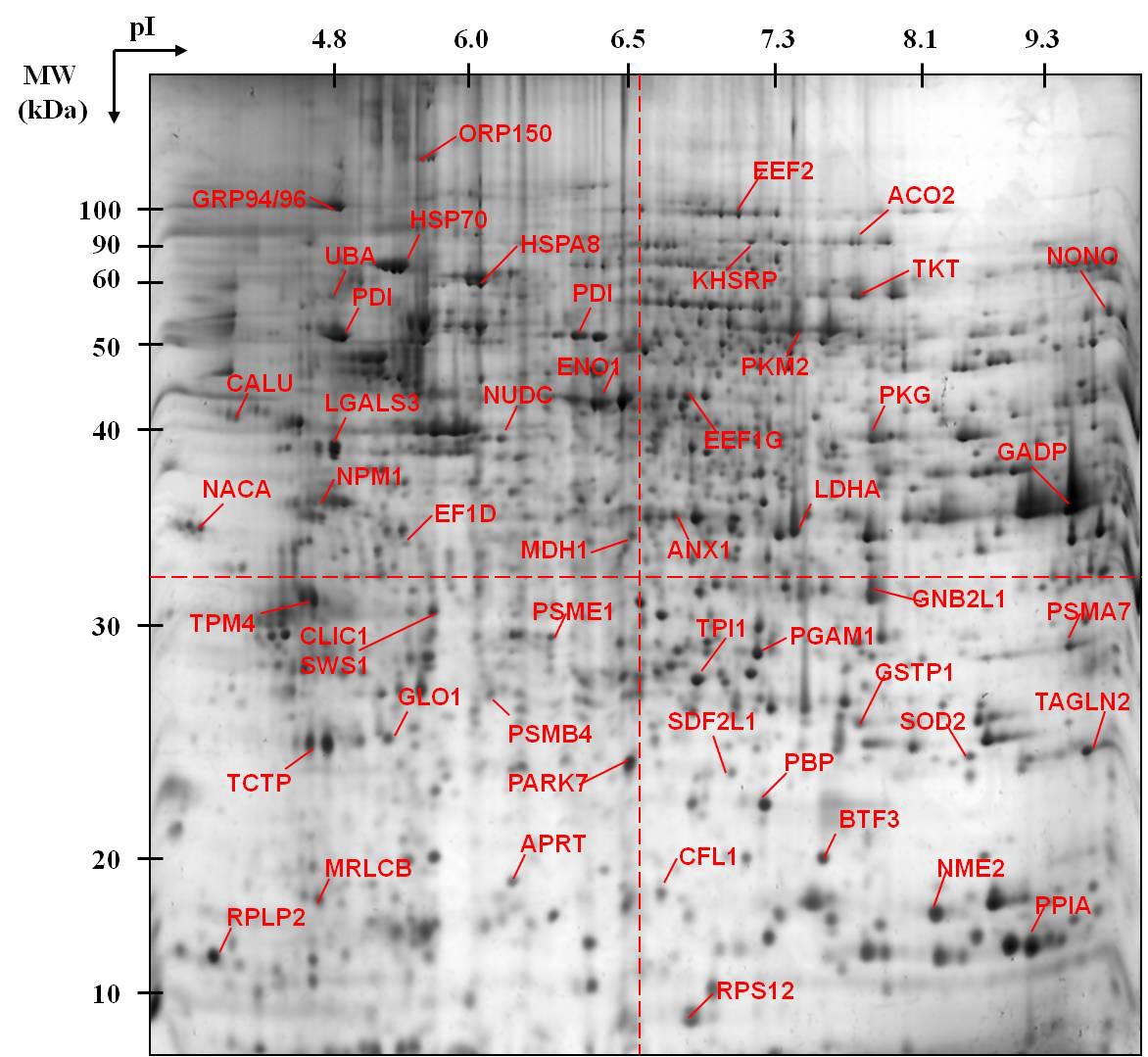
Experimental Details
CHO-K1 cells were grown in Ham’s F12 medium supplemented with 10% fetal bovine serum.
Samples were separated on a 3-10 linear pH gradient and 12%T polyacrylamide gel. Gels were stained with SYPRO Ruby and MALDI-TOF/TOF was used to identify spots.