News
Updated chromosome-scale CH assembly and CH/CHO-K1 annotations
January 27, 2021
A significantly more continuous version of the Chinese hamster genome, CH PICRH (GCF_003668045.3), along with its 2020 RefSeq annotation is now on CHOgenome.org. Long-range scaffolding of the previous CH genome assembly was performed using high-throughput chromosome conformation capture (Hi-C) and now 97% of the genome is contained in 11 large scaffolds corresponding to the CH chromosomes. In addition, the updated 2020 RefSeq annotation for the CHO-K1 cell line is available. Both can be searched (gene search and BLAST) and viewed on JBrowse. More information on the most recent version of the Chinese hamster genome can be found here.
Updated CH assembly and CH/CHO-K1 annotations
May 1, 2019
We are excited to announce that the significantly improved Chinese hamster genome, CH PICR (GCF_00366804.1), along with its 2018 RefSeq annotation is now online. In addition, an updated RefSeq annotation for the CHO-K1 cell line is now available. Both can be searched (gene search and BLAST) and viewed on JBrowse. More information on the updated Chinese hamster genome can be found here.
New mRNA Expression Browser (Beta)
June 7, 2017
The browser (beta version) for the visualization of mRNA expression from CHO-K1 cells is now online. The data is from several published DNA-microarray or RNA-Seq experiments. The tutorial on how to use this browser can be found here.
Tutorial 4 – BLAST Searching the CHO Genome
August 2014, Version 2.0
Click here for PDF version.
Accessing the CHO Genome BLAST Tool
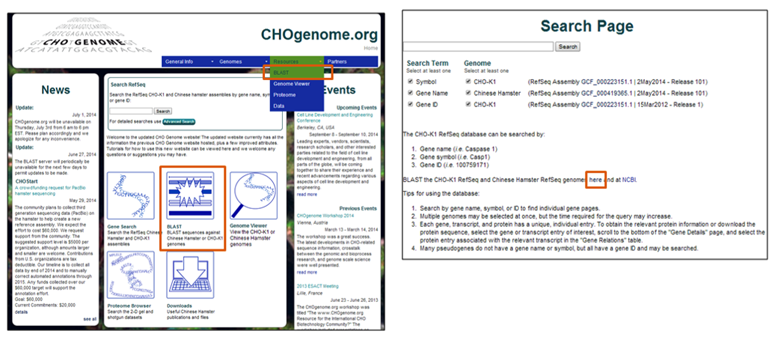
The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting "BLAST" from the menu bar under the Resources tab. This tab is available on all web pages within the CHO genome project. An additional link to the CHO genome BLAST web server is also provided on the CHO genome search pages, as well as a link to the NCBI BLAST web server.
Select the BLAST icon on the home Click on the BLAST server link on the CHO
page or from the resources tab genome search pages
Using the CHO Genome BLAST Tool
1) The CHO BLAST page allows for BLAST searches against the CHO and Chinese hamster (CH) genome databases.
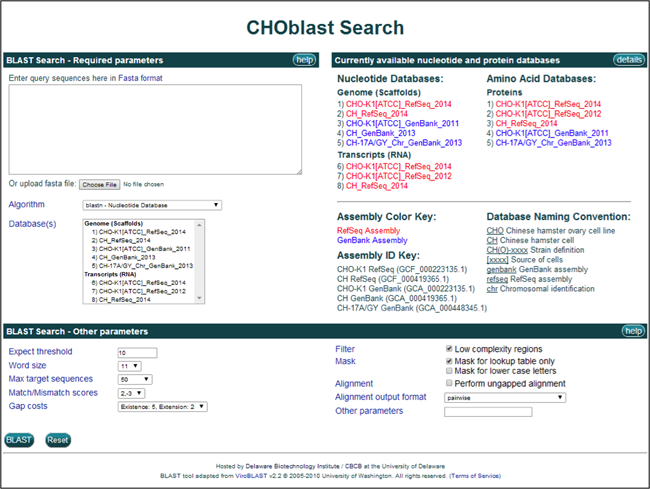
The nucleotide and amino acid databases hosted on the Chinese hamster genome database are listed to the right of the Basic Search panel. The nucleotide databases are divided into Genome (scaffold) and Transcript (RNA) databases, while the amino acid databases consist only of protein databases. The organism or cell line of origin is listed first, followed by the type of assembly (RefSeq or GenBank), and finally the year of release. The keys for the abbreviations and naming conventions are listed below these database lists.
2) Additional details regarding the multiple BLAST programs and databases are available. Clicking on the Algorithm link provides a brief description of the BLAST programs.

Clicking on the Database(s) link or the details button in the "Currently available nucleotide and protein databases" section title bar brings up a webpage with a brief description of the databases currently available for BLAST searching, including the name, version, date, and a link to the original publication article.

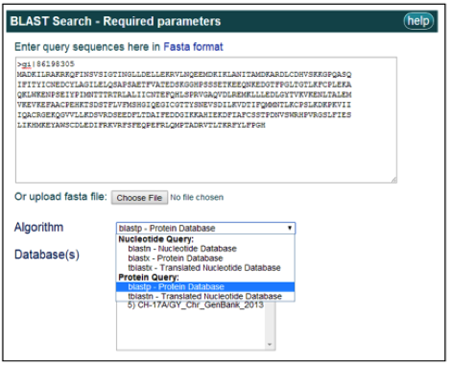 3) Query sequences in FASTA format can be pasted into the search box at the top of the page or uploaded as a FASTA file. Multiple query sequences may be entered for each search.
3) Query sequences in FASTA format can be pasted into the search box at the top of the page or uploaded as a FASTA file. Multiple query sequences may be entered for each search.
The BLAST program and database are then selected from the currently available options.
For example, to BLAST the most recent CH genome protein database, select the blastp program and the CH_RefSeq_2014 database from the "Proteins" list.
To perform a basic BLAST search, click the BLAST button after all the above information is entered and selected. If you wish to perform a more advanced search, do not hit basic search yet and proceed to instruction #4.
4) In the BLAST Search – Other parameters section, the default BLAST parameters can be varied to perform an altered, more advanced BLAST search.

Clicking on the highlighted blue terms (such as Expect threshold, etc.) will provide a brief description of each advanced search parameter that can be varied.
To perform an advanced BLAST search, click the BLAST button once all the required information is entered and the advanced parameters are altered.
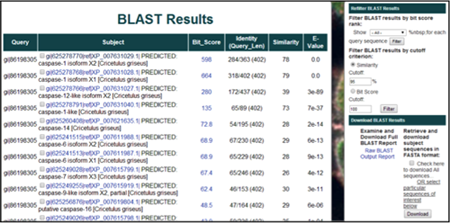 5) The results of the BLAST alignment are summarized in a table with the query sequence name, the subject sequence name, the bit score, the identity length, the identity percentage, and the E-value.
5) The results of the BLAST alignment are summarized in a table with the query sequence name, the subject sequence name, the bit score, the identity length, the identity percentage, and the E-value.
The results can be filtered by score (showing only the top 1, 5, or 10 alignments), by Similarity Cutoff Percentage, or by BLAST Bit Score.
After entering the filter parameter, click either the
"Filter" or the "Parse again" buttons to refresh the results table.
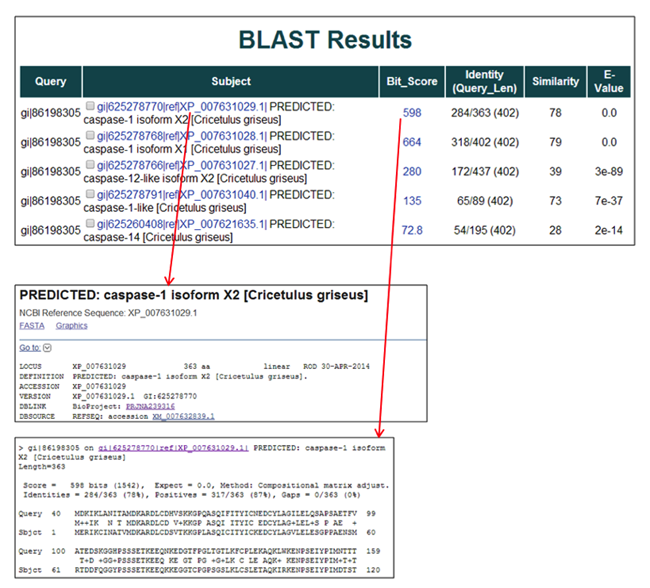
To view the RefSeq/GenBank entry for each subject sequence, click on the sequence name in the Subject column (i.e. gi|625278770|ref|XP_007631029.1|).
To view the pair-wise alignment for a specific alignment, click on the value in the Score column for any alignment (i.e. 598).
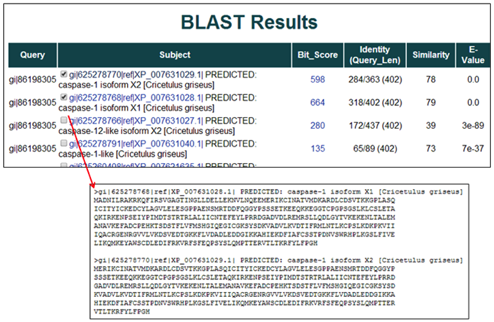
6) To download the resulting BLAST sequences, click the "Check here to download all sequences" box or select individual subject boxes within the table and then hit the "Download" button. A text file of the selected BLAST sequence(s) in the FASTA format will open in a new window.
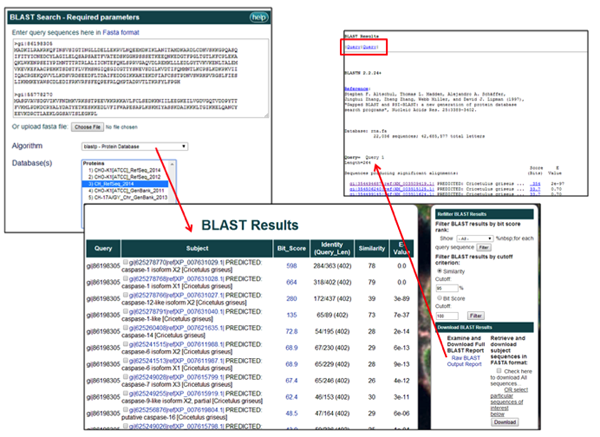
7) Multiple query sequences can be BLAST searched at one time. The results from all query sequences will be displayed in a single results table. To inspect all the BLAST pair-wise alignments, click on Raw BLAST Output Report, the link located under the "Download BLAST Results" heading. Links provided at the top of the page, in the red outlined box, can be used to quickly navigate between the alignment results for each, individual query sequence.
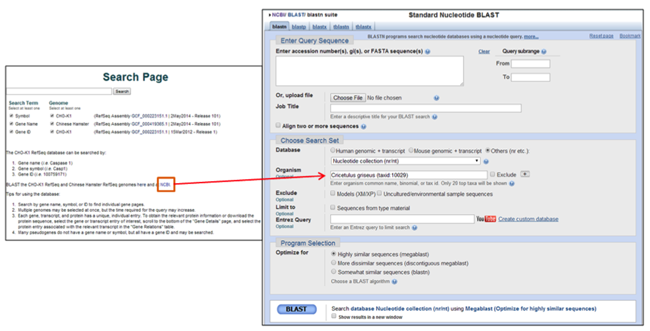
BLAST searching the CHO-K1 Genome at NCBI
A link to the NCBI BLAST web server is also provided on the CHO-K1 genome search pages. To BLAST the CHO genome using the NCBI BLAST web server, enter the required BLAST information and select the "Cricetulus griseus WGS" database under the "Choose Search Set" menu.