News
Updated chromosome-scale CH assembly and CH/CHO-K1 annotations
January 27, 2021
A significantly more continuous version of the Chinese hamster genome, CH PICRH (GCF_003668045.3), along with its 2020 RefSeq annotation is now on CHOgenome.org. Long-range scaffolding of the previous CH genome assembly was performed using high-throughput chromosome conformation capture (Hi-C) and now 97% of the genome is contained in 11 large scaffolds corresponding to the CH chromosomes. In addition, the updated 2020 RefSeq annotation for the CHO-K1 cell line is available. Both can be searched (gene search and BLAST) and viewed on JBrowse. More information on the most recent version of the Chinese hamster genome can be found here.
Updated CH assembly and CH/CHO-K1 annotations
May 1, 2019
We are excited to announce that the significantly improved Chinese hamster genome, CH PICR (GCF_00366804.1), along with its 2018 RefSeq annotation is now online. In addition, an updated RefSeq annotation for the CHO-K1 cell line is now available. Both can be searched (gene search and BLAST) and viewed on JBrowse. More information on the updated Chinese hamster genome can be found here.
New mRNA Expression Browser (Beta)
June 7, 2017
The browser (beta version) for the visualization of mRNA expression from CHO-K1 cells is now online. The data is from several published DNA-microarray or RNA-Seq experiments. The tutorial on how to use this browser can be found here.
Tutorial 6 – Searching the Proteome Database
August 2014, Version 2.0
Click here for PDF version.
The Chinese hamster proteome database can be reached by selecting the "Proteome" resource from the menu bar or by selecting the "Proteome Browser" button from the homepage.

The address for the Chinese hamster proteome database is http://www.CHOgenome.org/proteome.php.
The proteome database provides access to the proteomic data generated from CHO cell lines. This database hosts protein data from 2D polyacrylamide gel electrophoresis (2D PAGE) gels and shotgun proteomics analysis.
To have your data included, please contact us at chogenome@dbi.udel.edu.
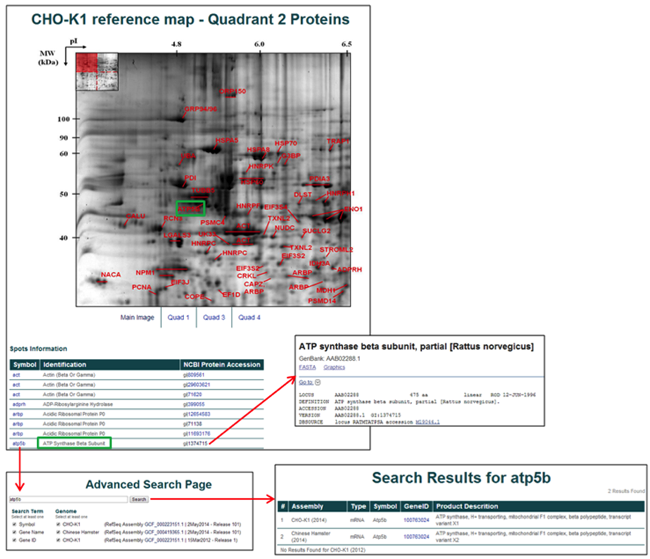
2D PAGE Database
The 2D PAGE database is accessed by selecting the 2D PAGE Results button.

The webpage has the complete list of 2D gels available for public search with a small image of the gel and the publication information, if it has been published.

Selecting an image leads to a much larger image of the gel with a link to the reference (if applicable).

Selecting a quadrant (if the total number of identified proteins is large) leads to a table of the identified proteins, a labeled 2D gel, and links to the NCBI protein and the CHO Genome gene pages.
Shotgun Proteomics Database
The shotgun proteomics database is accessed by selecting the Shotgun Results button.

The shotgun proteomics database consists of the protein sets submitted to the CHO genome website. The publication for each dataset (if applicable) is listed following the description of each dataset. The link connects to the Publications page on the CHO genome website, which contains the links for the relevant publications.
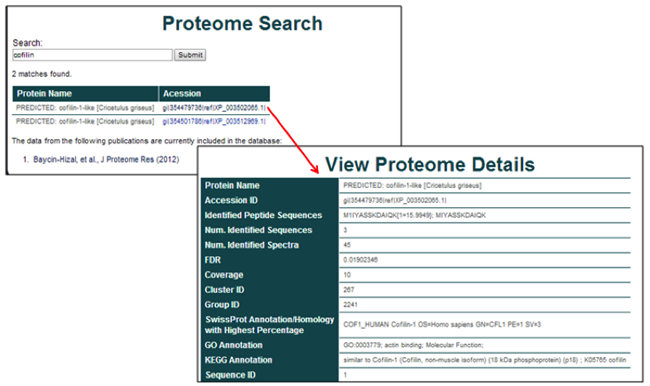
The shotgun proteome database is searchable by protein name. The results page consists of all matches from the selected datasets. The protein name and accession identification are provided for each result.
Selection of the accession identification yields the specific details from the identification. These details can consist of the protein name, accession ID, the peptide sequences that were identified, the number of identified sequences, coverage, FDR, and other experiment specific details of possible relevance, including the cluster ID, group ID, SwissProt Annotation Homology, GO annotation, and Sequence ID.